



|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
Our team is exploring how innovative analyses of big data sets can be used to discover new therapeutics. In one of our recent projects, we developed a new algorithm, TargetTranslator, which could identify new treatments for neuroblastoma (Almstedt et al, Nature Communications 2020). Our current work explores how data-driven methods can be used to target a broader range of pediatric cancer, including medulloblastoma and DIPG. Our team has invented several other tools for data-driven cancer research, such as CancerLandscapes, EPoC, and CoPIA.
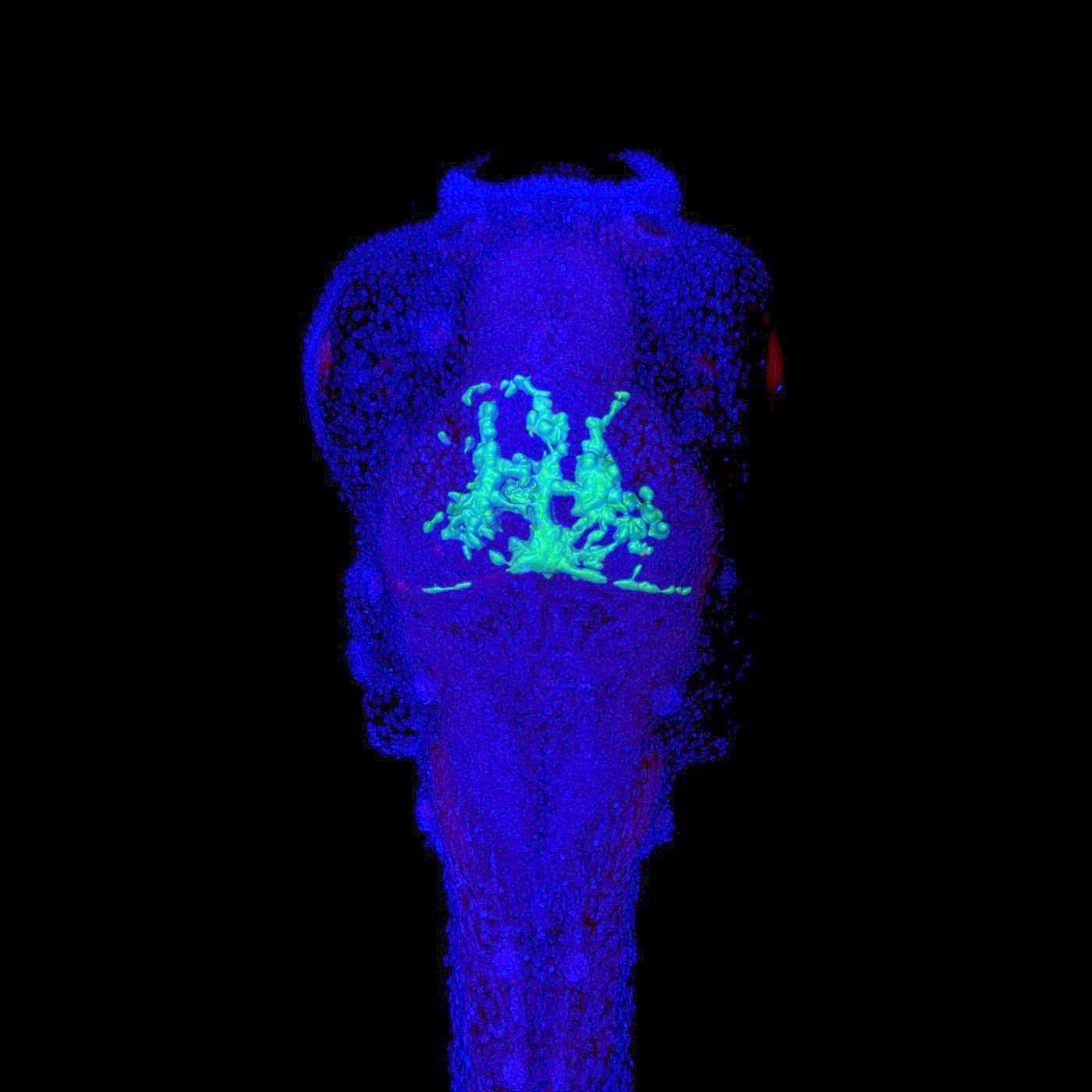
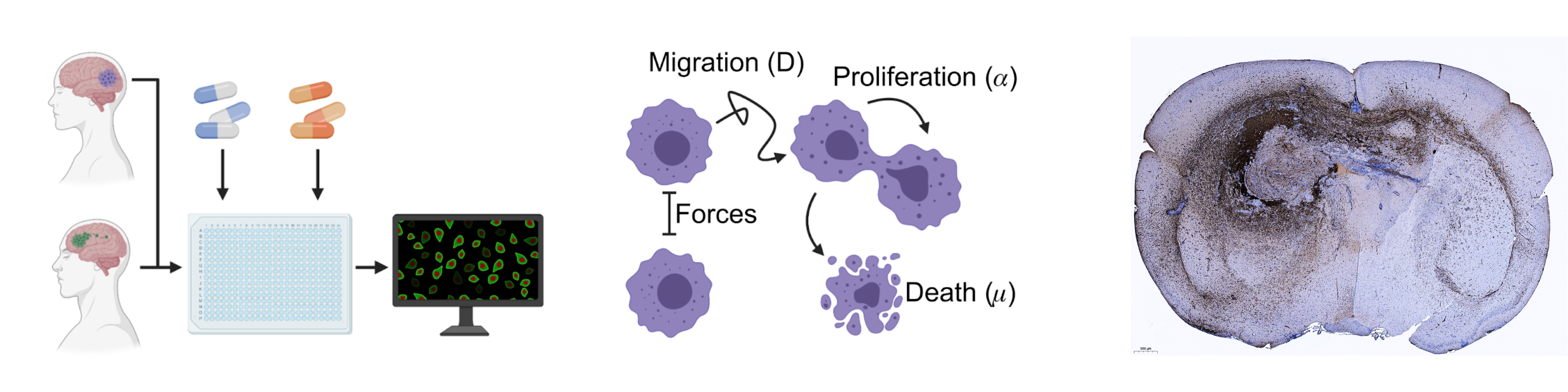
 Decoding adult brain tumor invasion and drug response.
Decoding adult brain tumor invasion and drug response.
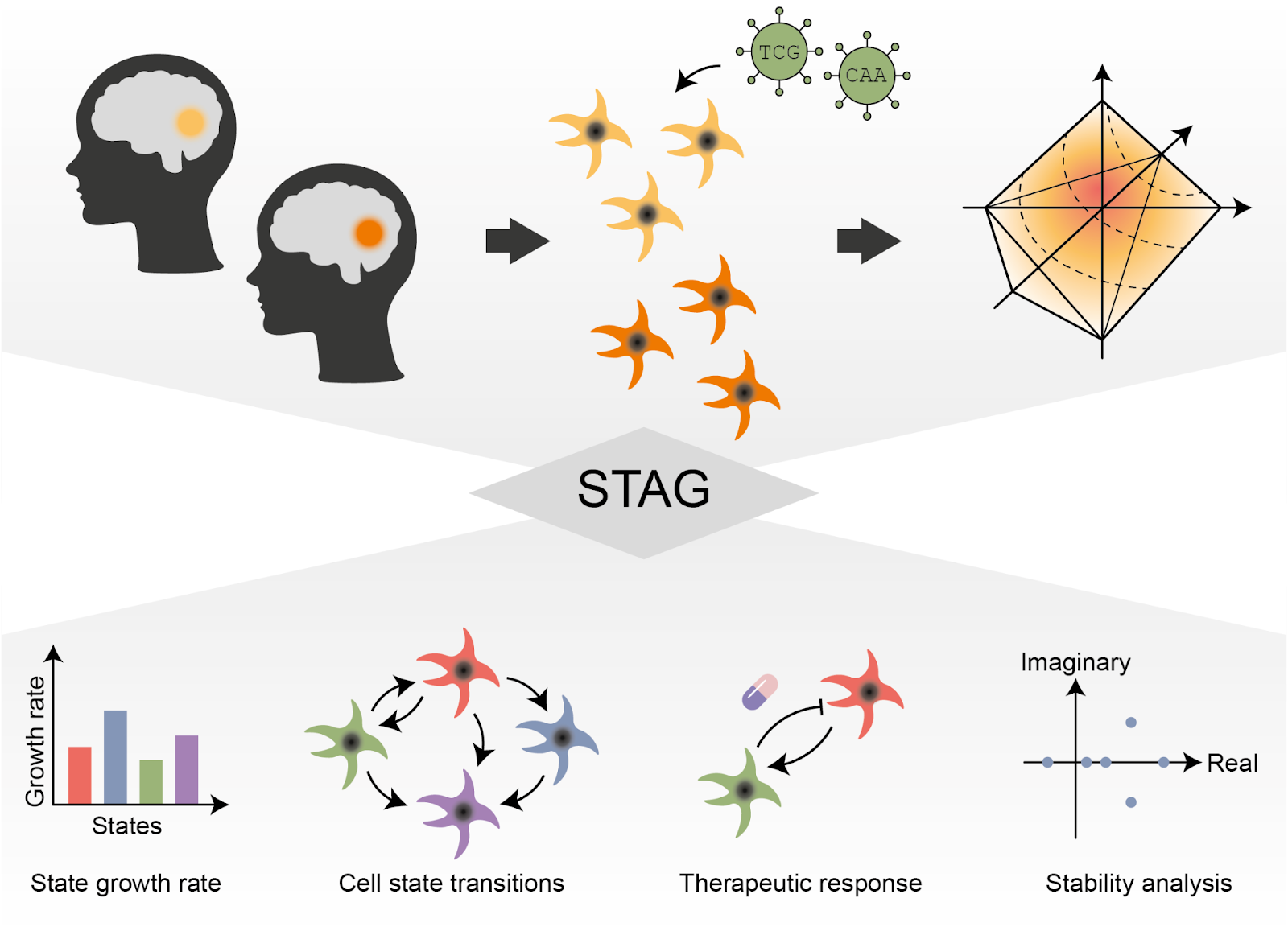 Brain tumor systems biology: controlling cell states
Brain tumor systems biology: controlling cell states